|
|
 |
Affy Analysis PipelineThe Affy Analysis Pipeline allows a user to group together Affy Arrays, perform normalization and find differentially expressed genes. In addition the data generated from finding differentially expressed genes can be easily added into SBEAMS which is then accessible via the GetExpression web page. Data queried via GetExpression can be directly loaded into Cytoscape. See below for more information about each of the steps
| Analysis help | | Use the image below to navigate to different analysis help pages | 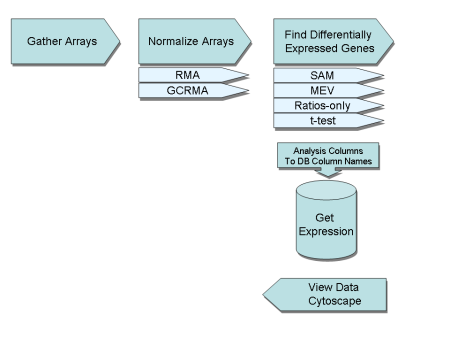 |
| Affy Analysis Pipeline Overview | | The Pipeline is broken down into three discrete parts which are Grouping Array Files, Normalization and Analysis. The analysis pages are all based on some CGI-software downloaded from the Bioconductor library called webbioc. Software was developed to try to fully integrate these pages into SBEAMS to help increase data flow and ease of use. |
| Group Affy Arrays | | The first step of the pipeline is to start a new session within a project and then gather together a group of arrays for analysis |
| Normalize Data | | Once all the files are grouped together, the file group can be moved onto a Normalization step which is composed of a number of steps. The process actually takes the group of CEL files and pre-process the data through (RMA or GCRMA) and then performs quantilie normalization to produce a file of normalized expression values. |
| Run Analysis | | Find differentially expressed genes by running either SAM or t-test based method. The SAM method is currently the default method, developed by Tusher et al. and has been shown to do a good job at detecting differentially expressed genes. |
| Add Data to GetExpression | | To add data to the GetExpression follow these steps |
| Query Data from GetExpression | | Query Data from GetExpression. This page will allow the data to be queried and filtered in a multitude of ways. Once the results sets is produced the data can be saved in a variety of ways and the data maybe loaded into Cytoscape. See below about launching Cytoscape with Affy expression data. |
| Cytoscape and Affy Expression Data | | Cytoscape can be automatically launched to view Affy expression data (other expression data types can be used too). This method can easily upload all the expression data for an experiment an allows the user to view which differentially expressed genes are shared between the different conditions. |
|

