|
|
 |
R vs. GCOS MAS5 ComparisonComparison of GCOS/MAS5.0 Output and Bioconductor/affy/mas5 Output. Evaluated on Mouse 430 2.0, HG-133 Plus 2.0 and YG_S98 chips.
| Getting GCOS/MAS5 data | | In GCOS, double-click names of .CHP files desired. Data is displayed in tabular format, choose File->Save to save it out. |
| Getting R/Bioconductor/affy/mas5 data | | Create a list file with a few chips of each chip type. The first line is the chip name, second line is always blank and starting with the third line are the CEL file names. Both the directory under Affymetrix/core/probe_data and the actual CEL file name must be included. |
| LPS_0_120.list | | Mouse430_2
200406/20040621_01_LPS2-0.CEL
200406/20040622_01_LPS3-0.CEL
200406/20040621_06_LPS2-120.CEL
200406/20040622_06_LPS3-120.CEL |
| YG_S98-test.list | | YG_S98
200409/20040920_01_G848I.CEL
200409/20040920_02_G848II.CEL
200409/20040922_03_G848III.CEL
200408/20040831_05_G984II.CEL
200409/20040916_02_G984II.CEL
200408/20040831_06_G984III.CEL |
| hgu133plus2-test.list | | /net/arrays/Affymetrix/core/probe_data/200404/20040421_01_LN_1.CEL
/net/arrays/Affymetrix/core/probe_data/200404/20040421_02_LN_2.CEL
/net/arrays/Affymetrix/core/probe_data/200404/20040421_03_C4-2_2.CEL |
| Bioconductor Steps | For each chip type, use R/Bioconductor
| 1) | R can be downloaded from
| |
| 2) | Bioconductor packages can be installed by running within R:
| |
|
source("http://www.bioconductor.org/getBioC.R")
getBioC()
| | 3) | Then run in R
| |
| file.table.name <- “<list file name>”
output.file.name <- “<list name>.txt”
switch(.Platform$OS.type, unix = .libPaths("/net/arrays/Affymetrix/bioconductor/library/"),"windows")
require(affy)
ft <- read.table(file=file.table.name,sep="\t",blank.lines.skip=FALSE)
ft.character <- c(as.vector(ft$V1))
setwd(switch(.Platform$OS.type, windows = "Z:/Affymetrix/core/probe_data","/net/arrays/Affymetrix/core/probe_data"))
data <- read.affybatch( filenames = c(ft.character[ 3:length(ft.character) ]) )
eset <- mas5(data,sc=250)
PACalls <- mas5calls(data)
Matrix <- exprs(eset)
output <- cbind(row.names(Matrix),Matrix,se.exprs(PACalls))
write.table(output,file=output.file.name,sep="\t",row.names=FALSE)
|
|
| Comparison of Signal Values | | Signal values between R and GCOS show Pearson correlation of approximately 1, and slope very near 1. | 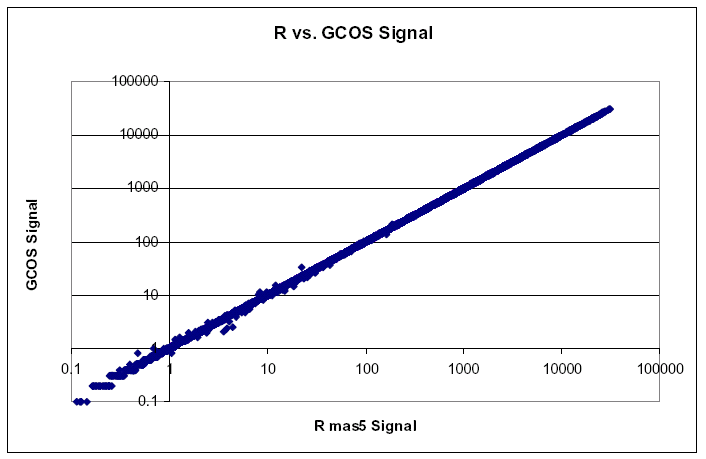 |
| Comparison of Detection p-values | | Detection p-values show a Pearson correlation near 1, however there are large differences in the low p-values that are masked by good correlation of large p-values | 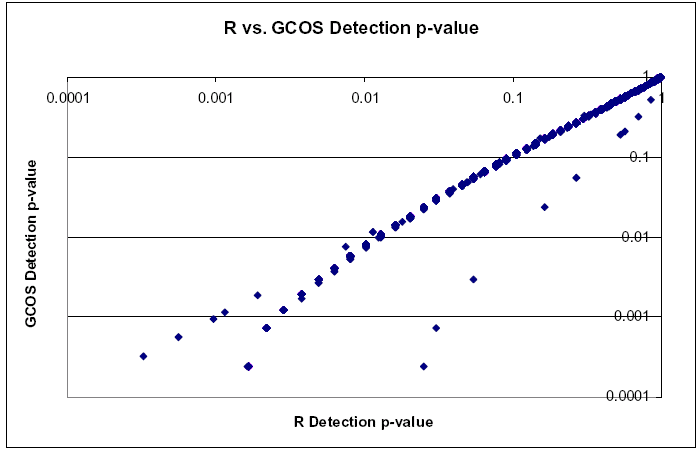 |
| Comparison of Detection Calls | |
Fortunately, differences in Detection p-values are not of great concern since it is the Detection Call that is actually used. For detection calls, Affy currently says p < 0.05 is P(present), 0.05 < p < 0.065 is M(marginal) and p > 0.065 is A(absent). Using these cutoffs, 4 Mouse 430 2.0 and 3 Hg-U133 Plus 2.0 gave a total of only 45/344429(0.013% difference) calls incorrect due to differences between R and GCOS. The reason there are any discrepancies at all is that while Affymetrix has disclosed their method for making detection calls, implementing the method as they describe it does not reproduce the results GCOS produces. |
|


