|
OverviewIn order to better support the data being generated by local microarray, proteomics, macroarray, and other experiments, we are developing and using SBEAMS (Systems Biology Experiment Analysis Management System), a framework for collecting, storing, and accessing data produced by these and other experiments. There is currently a significant amount of effort in the community to determine standards for storing microarray data (MAGE-OM/ML, GeneX, ArrayExpress, SMD, etc; see below), and as such we are working with these emerging standards so that we may leverage this effort in our own work. This will allow the data produced at ISB to be easily exported and will facilitate our efforts to correlate our own experimental results with external datasets. SBEAMS is modular in design to allow the storage of various types of experiments in the system; the different experiments can be managed separately but then correlated later under the same framework. Currently, development is being driven by the Microarray, Proteomics, and Inkjet Array projects, but additional experiments will be added. One of the goals of SBEAMS is to allow a user to request a set of experiments through a web interface from an ISB core facility, and be informed when the fully processed data are available in the database for subsequent analysis and annotation. The process of data acquisition, from sample preparation, spotting, hybridization, quantitation, and derivation of expression measures for microarray experiments as well as sample preparation, MALDI plate spotting, mass spectrometry, sequence database searches, and annotation for Proteomics experiments will be tracked by the "Systems Biology Experiment Analysis Management System" (SBEAMS) data management package under development at ISB. This integrated system is a consistent framework that combines a unified state-of-the-art relational database management system (RDBMS) back end, a collection of tools to store, manage, and query experiment information and results in the RDBMS, a web front end for querying the database and providing integrated access to remote data sources, and an interface to existing programs for clustering and other analysis. Since all data from each step of the experiment are warehoused in a modular schema in the RDBMS, quality control and data analysis tasks are greatly simplified. Within the SBEAMS framework, each investigator may first store and manage the data unique to his or her experiment. Then, the parameters of the process of executing the microarray, proteomics, or other experiment are captured into the database. The experimental data products are loaded into the database and an automated pipeline processes the raw data into gene expression measures with data quality estimates or protein matches and quality scores. The investigator may then use the SBEAMS built-in tools or custom scripts built on top of the framework to correlate the experimental results and experiment conditions and further understand the experimental results. Investigator annotations are also captured in the database for later analysis and correlation with other experiments. The SBEAMS-Microarray module will be compliant with the emerging MAGE-OM/ML (MicroArray Gene Expression - Object Model/Markup Language) specification (http://sourceforge.net/projects/mged/), a work-in-progress which will combine all previous standards such as MAML, GEML, GeneXML, etc. The ISB is actively contributing to this new standard. Our tools will be able to export all the resulting microarray datasets in the MAGE-ML format (once specification is complete), which will allow the data to be easily accessed by anyone in the community with MAGE-compliant tools. In addition, the SBEAMS will allow Internet access to the data via a public web front end when they are fully processed and released by the investigators. We are a member of the MGED (Microarray Gene Expression Database) Group (http://www.mged.org) and are committed to contributing to and using open standards like MIAME (Minimum Information About a Microarray Experiment) (http://www.mged.org/Annotations-wg/) and MAGE-ML for data exchange and storage formats, which will allow interoperability with other software like GeneX (http://genex.ncgr.org/), ArrayExpress (http://www.ebi.ac.uk/arrayexpress/), ArrayDB (http://genome.nhgri.nih.gov/arraydb/), ExpressDB (http://arep.med.harvard.edu/ExpressDB/ ), Stanford Microarray Database (http://genome-www4.stanford.edu/MicroArray/SMD/ ), etc. A few implementation details:SBEAMS is currently implemented in Perl using the Perl DBI module for database connectivity. This provides a good level of database back-end independence. ISB's principal back-end for SBEAMS is currently Microsoft SQL Server, but portions of SBEAMS are known to work with both MySQL and PostgreSQL. Sybase would also be viable due to high compatibility with MS SQL Server. Due to our collaboration with IBM, we will also adjust to support DB2 UDB as the back end; this is expected to be little trouble. The Perl modules that make up SBEAMS can be accessed by Perl CGI scripts which use the SBEAMS API to create a Web interface, and very similar (or often the same) Perl scripts can be executed from the command line or as part of automated batch jobs. Both web and command-line interfaces use a unified Authenticate() method which uses HTTP cookies for authentication if executed via a web server or UNIX login authentication if executed from the command line. The SBEAMS core module handles such tasks as user authentication, work group management, permissions management, simplified engine-independent SQL database access API, web form abstraction, tabular data rendering, and much more. One or more additional (experiment/project specific) SBEAMS modules are then invoked after the core module. These modules provide specific functionality to manage and browse microarray, proteomics, etc. experiments. At present, the Perl CGI's run on the Apache Web server http://db/ and the interactive Perl programs can run on interactive sessions on db or any other Linux/UNIX machine with the appropriate software installed. Certain components of SBEAMS have been written to assume a UNIX-like file structure and Perl implementation. It would likely be quite easy to modify SBEAMS to work equally well on any other operating system (e.g., Windows 2000) that can run Perl CGI scripts and Perl commands, but this has not been attempted .Screenshots and diagrams:
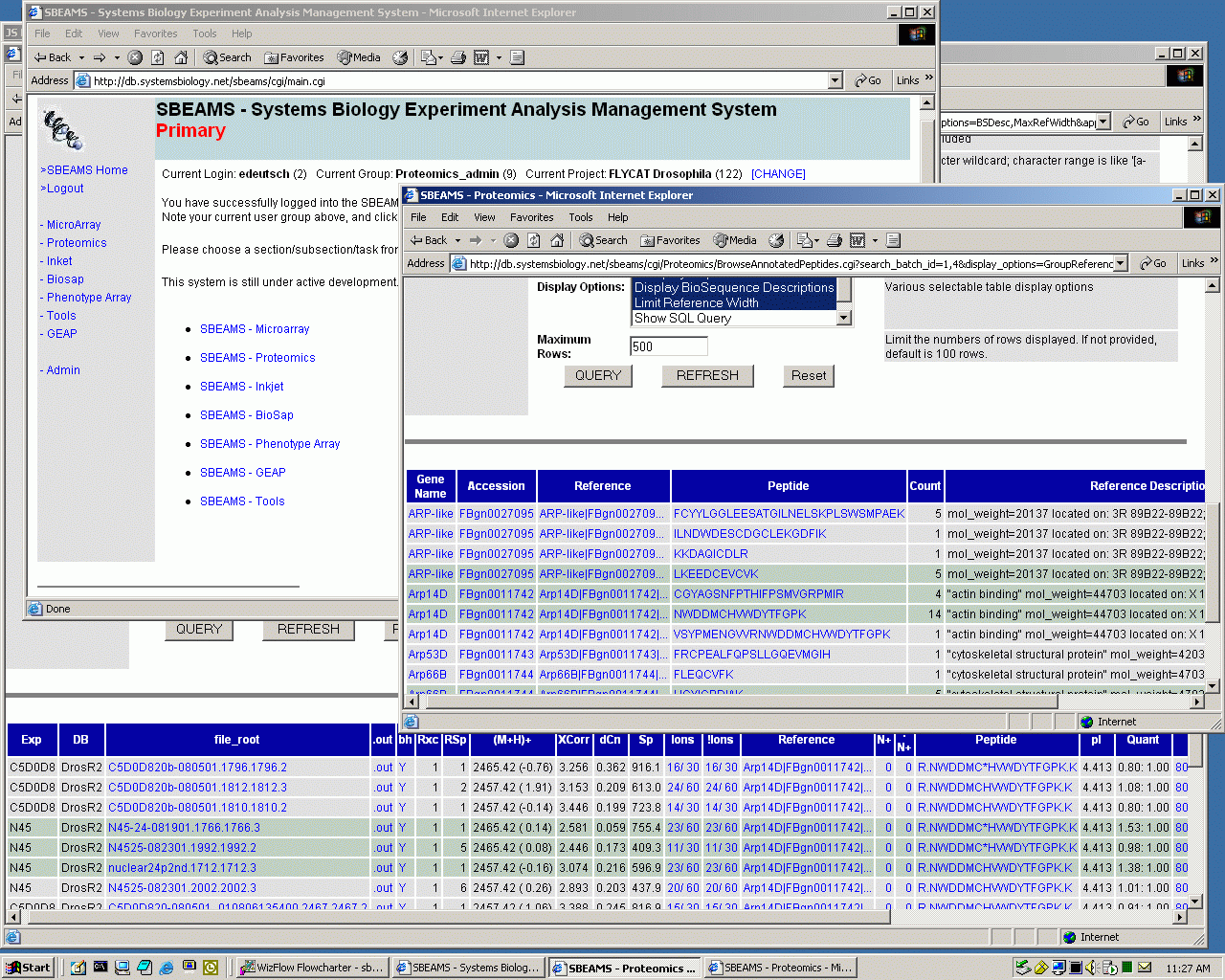
Below that to the right is a window in which the user has selected to issue a SQL query (query parameter entry fields are scrolled out of view) that summarizes the peptides that have been annotated for the "ARP*" genes in two Drosophila Proteomics experiments (click here to execute this query on the database - SBEAMS login required). Many peptides have been observed and annotated just once, while several have been annotated many times. Various hyperlinks give the user access to more information about the genes, proteins, and peptides actually observed and annotated. At the bottom is additional information about all the annotated occurrences of one of the peptides as identified by SEQUEST. The table includes information about which of the two experiments the peptides were observed in, the masses, the actual peptides (some of which contain tagged cysteines), pI values, ICAT quantitation ratios, annotation information (clipped off right edge) and much more. Search results can be annotated to additional insights. ISB Server/Data Access Configuration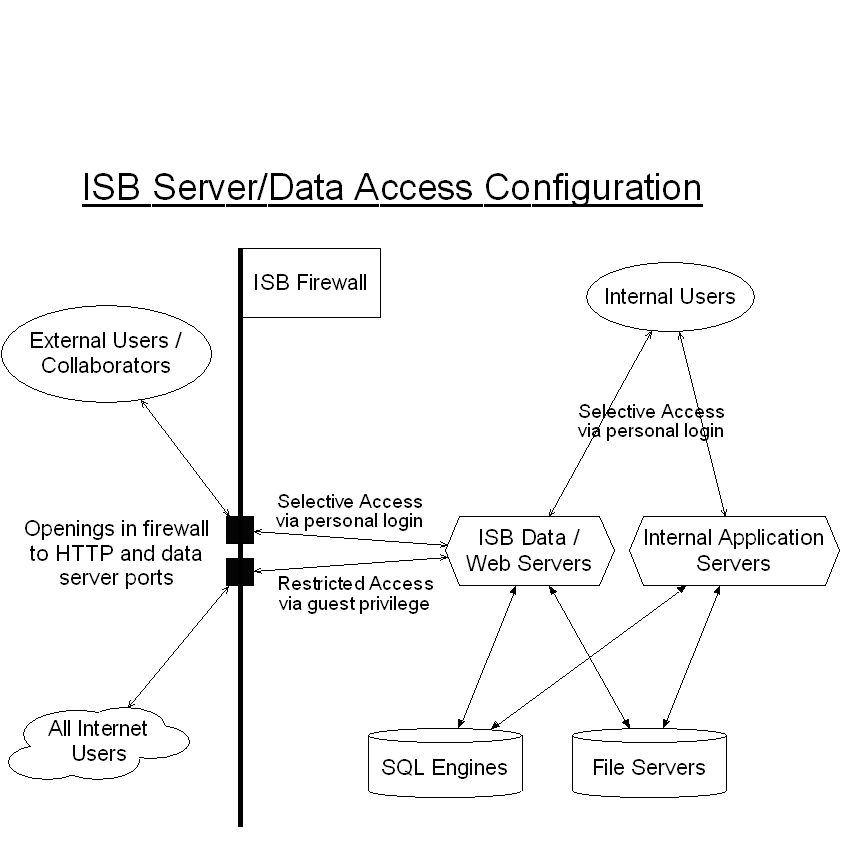
SBEAMS Data Access Configuration 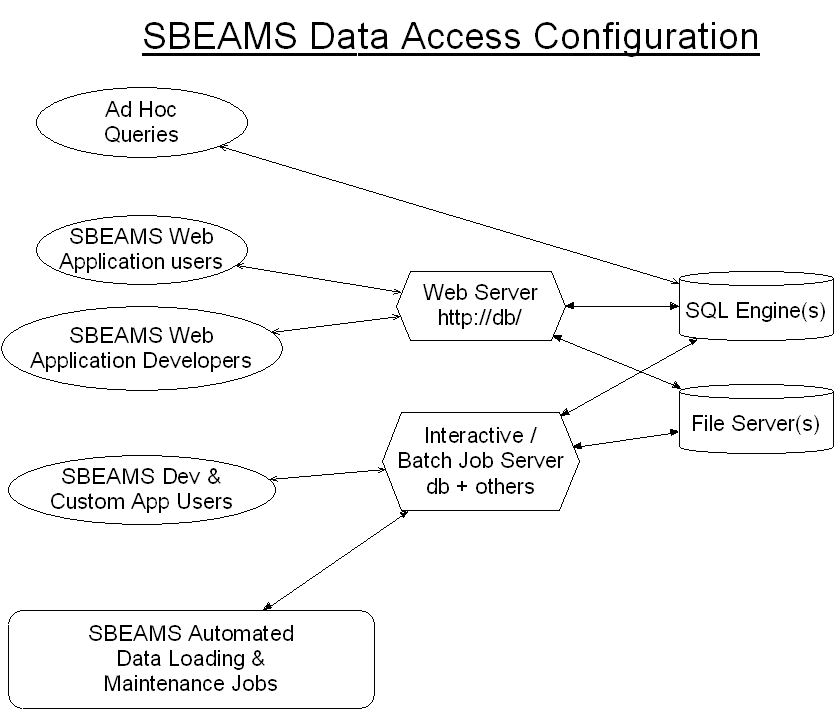
SBEAMS Modular Structure 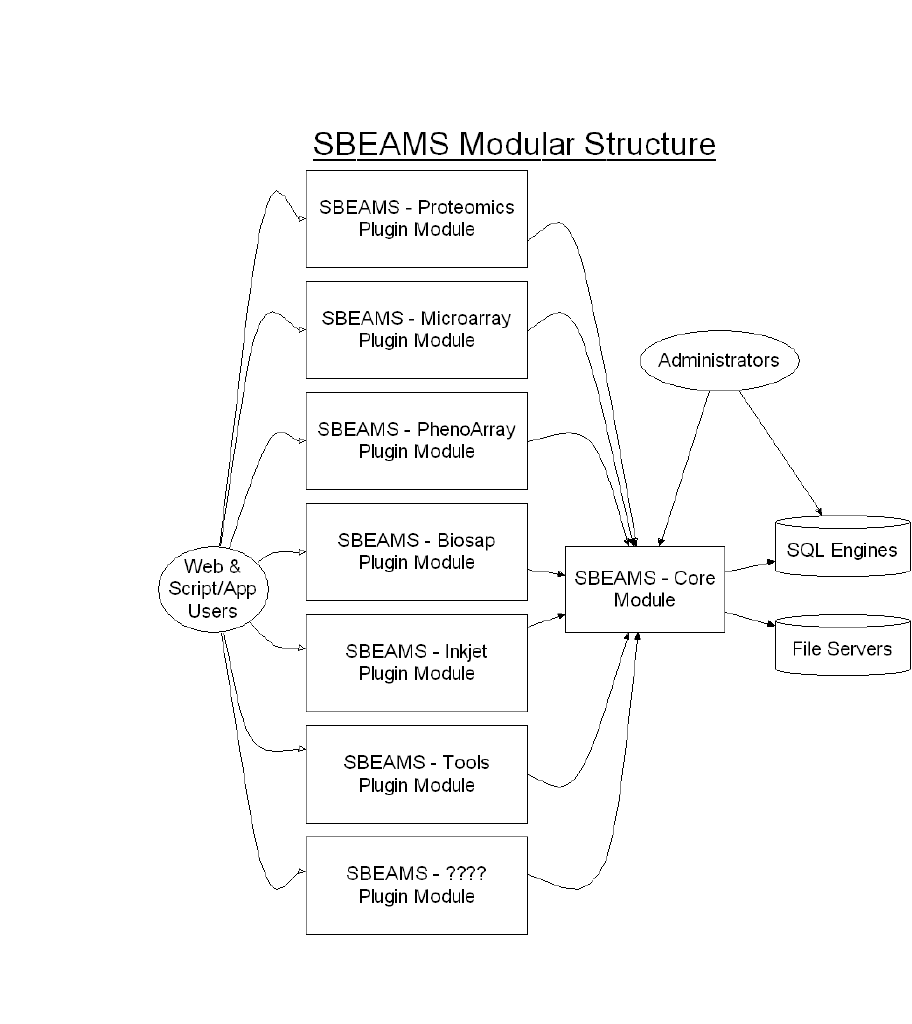
| |||||||